'Model-less' models
The model infrastructure can be used to read DICOM for 'noisy' labeling and to do pixel analysis and traditional computer vision labeling. Outputs are model prediction labels.
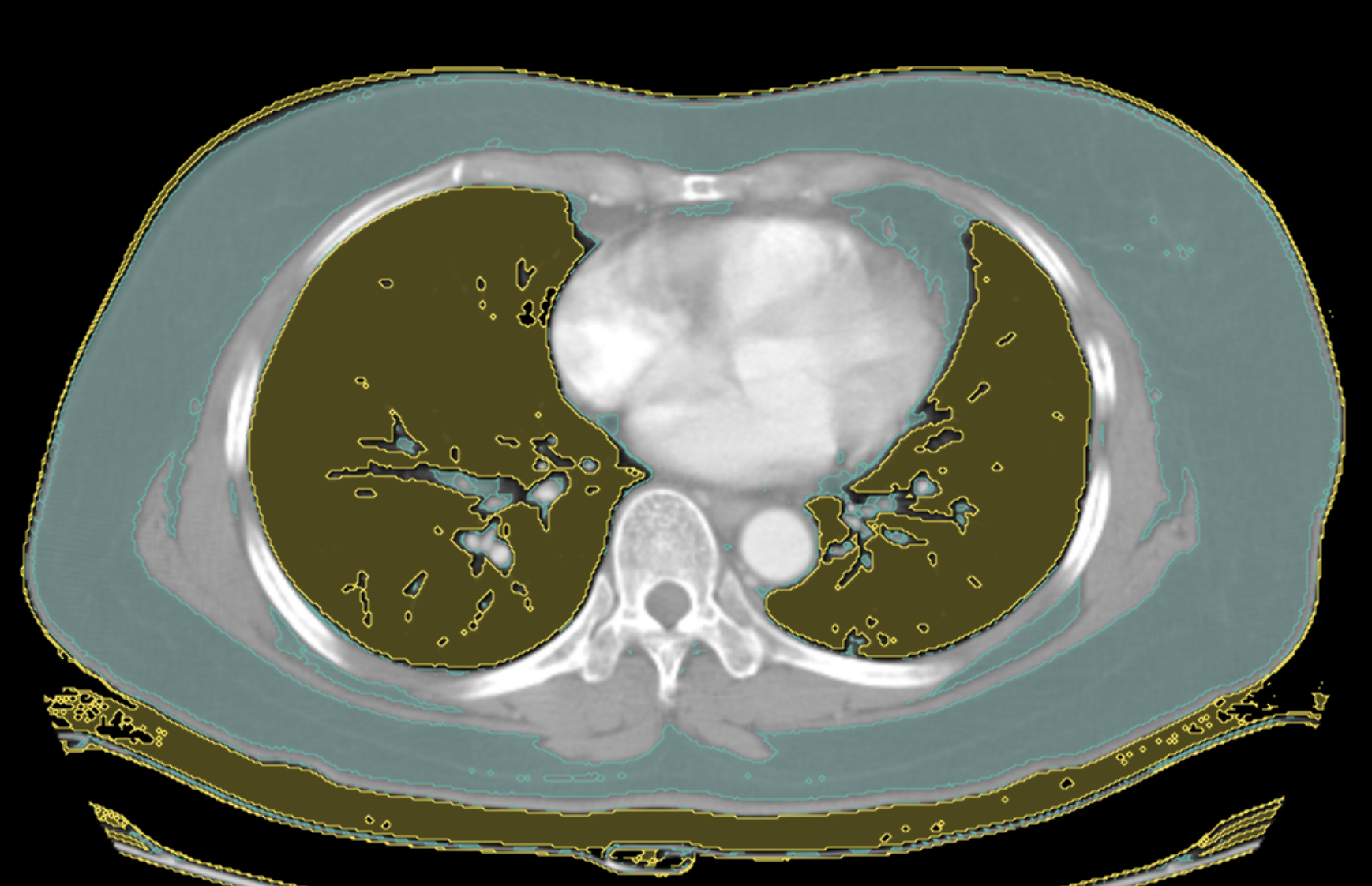
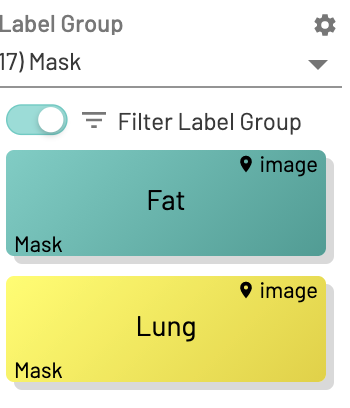
Mask label outputs:
Create 2 mask labels in the UI, one for Fat and one for Lung.
Below is some sample code which reads the DICOM tags to check modality and then creates fat and lung masks on the pixel array. Arguments for the minimum and maximum have default values but can be adjusted when the model is run. Outputs can be any type of label such as boxes, freeform shapes, global labels, etc
from io import BytesIO
import pydicom
from pydicom.pixel_data_handlers.util import apply_modality_lut
class MDAIModel:
def __init__(self):
self.model = None
def default_output(self, ds):
output = {
"type": "NONE",
"study_uid": str(ds.StudyInstanceUID),
"series_uid": str(ds.SeriesInstanceUID),
"instance_uid": str(ds.SOPInstanceUID),
}
return output
def predict(self, data):
input_files = data["files"]
outputs = []
for file in input_files:
if file["content_type"] != "application/dicom":
continue
ds = pydicom.dcmread(BytesIO(file["content"]))
if "Modality" in ds and ds.Modality != "CT":
continue
try:
array = apply_modality_lut(ds.pixel_array, ds)
except:
continue
# mask
# Load the input data arguments (if any)
input_args = data["args"]
# fat
mask = (array > int(input_args['Fat Min'])) & (array < int(input_args['Fat Max']))
outputs.append(
{
"type": "ANNOTATION",
"study_uid": str(ds.StudyInstanceUID),
"series_uid": str(ds.SeriesInstanceUID),
"instance_uid": str(ds.SOPInstanceUID),
"class_index": 0,
"data": {"mask": mask.tolist()}
}
)
# lung
mask = (array > int(input_args['Lung Min'])) & (array < int(input_args['Lung Max']))
outputs.append(
{
"type": "ANNOTATION",
"study_uid": str(ds.StudyInstanceUID),
"series_uid": str(ds.SeriesInstanceUID),
"instance_uid": str(ds.SOPInstanceUID),
"class_index": 1,
"data": {"mask": mask.tolist()}
}
)
return outputs