Import Annotations
1. Create MD.ai Client
We can use the mdai python client to quickly import a list of annotations into a project. For example, we may wish to load the output results of a machine learning model as annotations, or populate project metadata labels with report or clinical data.
Import the mdai library and create a client mdai_client similar to the "Getting Started" section. Ensure the correct domain and provide your access token.
import mdai
# Get variables from project info tab and user settings
DOMAIN = 'public.md.ai'
YOUR_PERSONAL_TOKEN = 'a1s2d3f4g4h5h59797kllh8vk'
mdai_client = mdai.Client(domain=DOMAIN, access_token=YOUR_PERSONAL_TOKEN)
We will need the project ID and dataset ID for import, click on the "Info" button in the bottom left sidebar in the UI. When more than one dataset is available within the project, select one dataset for import at a time.
Example IDs:
2. Create labels
If you are not importing to previously created labels, you'll need to create them in the UI and then get label IDs:
- In UI, use
Label ControlsandShow label ID, then copy and paste label IDs into your notebook - or download annotation JSON file and extract label dataframe using library and create a dictionary of labelnames and IDs
Example
# Replace with your filename
JSON = 'mdai_public_project_MwBe19Br_annotations_labelgroup_all_2020-09-23-214038.json'
results = mdai.common_utils.json_to_dataframe(JSON)
# Labels
labels_df = results['labels']
# Label dictionary
labels = dict(zip(labels_df.labelName, labels_df.labelId))
3. Choose correct format
Annotations format
The annotations variable must be a list of dictionaries with the set of required fields dependent on the different label types. All annotations must contain a corresponding labelId.
- For exam-scoped labels,
StudyInstanceUIDis required. - For series-scoped labels,
SeriesInstanceUIDis required. - For image-scoped labels,
SOPInstanceUIDis required. - A
notefield can be added to any annotation. - For local image-level labels, an additional field
datais required, with the format ofdataspecified based on the example below. - For image-scope labels or local image-level labels for the specific frame of multi-frame data,
frameNumber(0-indexed) is required.
Pick the format that matches the annotations you are importing and convert your data to create a list of dictionaries.
Exam label
{
'labelId': 'L_jZRPN2',
'StudyInstanceUID': '1.2.276.0.7230010.3.1.2.8323329.19529.1513659878.548505',
'note': 'This is an annotation note that can be imported as well'
}
Series label
{
'labelId': 'L_nZYWNg',
'SeriesInstanceUID': '1.2.276.0.7230010.3.1.3.8323329.19529.1513659878.548504'
}
Image label (global label)
{
'labelId': 'L_3EMmEY',
'SOPInstanceUID': '1.2.276.0.7230010.3.1.4.8323329.19529.1513659878.548506'
}
Multi-Frame (global label)
{
'labelId': 'L_3EMmEY',
'SOPInstanceUID': '1.2.276.0.7230010.3.1.4.8323329.19529.1513659878.548506',
'frameNumber': 0,
}
Bounding box
{
'labelId': 'L_xZgpEM',
'SOPInstanceUID': '1.2.276.0.7230010.3.1.4.8323329.25053.1513659982.870619',
'data': {'x': 200, 'y': 200, 'width': 200, 'height': 400},
'note': 'Any label can have a note'
}
Polygon, freeform, or line
{
'labelId': 'L_nEK9AV',
'SOPInstanceUID': '1.2.276.0.7230010.3.1.4.8323329.14861.1513659866.150292',
'data': {'vertices': [[300,300],[500,500],[600,700],[200,800]]},
'note': 'Optional note'
}
Mask
Convert the mask numpy_arr to data for import by using the library funcion convert_mask_data
{
'labelId': 'L_nEK9AV',
'SOPInstanceUID': '1.2.276.0.7230010.3.1.4.8323329.14861.1513659866.150292',
'data': mdai.common_utils.convert_mask_data(numpy_arr),
'note': 'Optional note'
}
Location
{
'labelId': 'L_BA3JN0',
'SOPInstanceUID': '1.2.276.0.7230010.3.1.4.8323329.2010.1513660008.830854',
'data': {'x': 400, 'y': 400}
}
Example annotation list for bounding box
# List of annotation dictionaries
annotations = [
{
'labelId': 'L_xZgpEM',
'SOPInstanceUID': '1.2.276.0.7230010.3.1.4.8323329.25053.1513659982.870619',
'data': {'x': 200, 'y': 200, 'width': 200, 'height': 400},
'note': 'Optional note'
},
{
'labelId': 'L_eUwwYf',
'SOPInstanceUID': '1.2.276.0.7230010.3.1.4.8323329.25053.1513659982.870720',
'data': {'x': 34, 'y': 45, 'width': 120, 'height': 352}
},
#...
{
'labelId': 'L_xZgpEM',
'SOPInstanceUID': '1.2.276.0.7230010.3.1.4.8323329.25053.1513659982.871892',
'data': {'x': 233, 'y': 144, 'width': 26, 'height': 38}
}
]
Import the annotations array you just created
Example output:
Importing 8 annotations into project LxR6zdR2, dataset D_vBnWb1.
Successfully imported 8 / 8 annotations into project LxR6zdR2, dataset D_vBnWb1.
Overall import errors will be printed to the console. However, even with a successful import, there may be errors with importing individual annotations: these are returned as a list.
failed_annotations = mdai_client.import_annotations(annotations, project_id, dataset_id)
print(failed_annotations)
# [{'index': 0, 'reason': 'Invalid labelId L_zzzzzz'}, {'index': 1, 'reason': 'Invalid labelId L_zzzzzz'}]
Metadata
Metadata labels are imported in exactly the same way as but are first created in the Metadata label group section in the UI
metadata_annotations = [
# metadata label (exam-scoped)
{
'labelId': 'L_7E5pMZ',
'StudyInstanceUID': '1.2.276.0.7230010.3.1.2.8323329.19529.1513659878.548505',
'note': 'Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Enim sed faucibus turpis in eu ipsum suspendisse ultrices gravida. Praesent elementum facilisis leo vel. Donec enim diam vulputate ut pharetra sit. Vel pretium lectus quam id leo in vitae '
},
# metadata label (series-scoped)
{
'labelId': 'L_pEnelZ',
'SeriesInstanceUID': '1.2.276.0.7230010.3.1.3.8323329.19529.1513659878.548504',
'note': 'Information to be imported'
},
# metadata label (image-scoped)
{
'labelId': 'L_yZBWwE',
'SOPInstanceUID': '1.2.276.0.7230010.3.1.4.8323329.19529.1513659878.548506',
'note': 'Information to be imported'
},
# list of additional annotations to import
#...
]
Import the metadata annotations
Example output:
Importing 3 annotations into project LxR6zdR2, dataset D_vBnWb1...
Successfully imported annotations into project LxR6zdR2.
Sublabels / child annotations
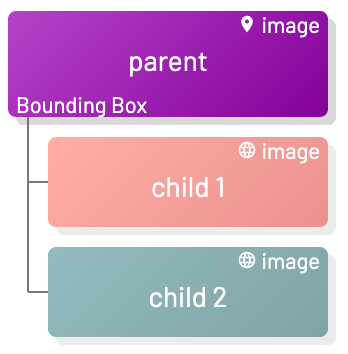
To import annotations for labels with sublabels, we can specify the sublabels using the children field. Each item in children should follow the appropriate format of its label type (see "Annotation format" section above). Ensure the specified DICOM resource UIDs match up with the label scopes.
The labelId of the children must be sublabels. In the following example, L_rEGG7E and L_mEzplA are sublabels of the label L_5AJeMN.
annotations = [
{
'labelId': 'L_5AJeMN',
'SOPInstanceUID': '1.2.276.0.7230010.3.1.4.8323329.31185.1513659797.320640',
'data': {'x': 200, 'y': 200, 'width': 200, 'height': 400},
'children': [
{
'labelId': 'L_rEGG7E',
'SOPInstanceUID': '1.2.276.0.7230010.3.1.4.8323329.31185.1513659797.320640',
},
{
'labelId': 'L_mEzplA',
'SOPInstanceUID': '1.2.276.0.7230010.3.1.4.8323329.31185.1513659797.320640',
},
]
},
# list of additional annotations to import
#...
]
Preserving original users / timestamps
Optionally, for each annotation, createdById / updatedById / createdAt / updatedAt may be included. For example, one may wish to import exported annotations from another project into a new project, while preserving the original user and timestamp information. For this to work properly, the specified createdById / updatedById must also be project users of the target project.
By default, when createdById / updatedById are not specified, the new annotations will be associated with the importing user.
Example:
annotations = [
{
'labelId': 'L_3EMGXA',
'StudyInstanceUID': '1.2.276.0.7230010.3.1.2.8323329.5420.1513659928.902121',
'createdById': 'U_eMzVMx',
'updatedById': 'U_eMzVMx',
'createdAt': '2023-09-20T01:27:44.016Z',
'updatedAt': '2023-09-20T01:27:44.026Z',
}
]
4. Import function
Example output:
Importing 3 annotations into project LxR6zdR2, dataset D_vBnWb1...
Successfully imported annotations into project LxR6zdR2.
(Optional) Import DICOM-SR as MD.ai Annotation or JSON
MD.ai Python client supports import_sr()function which is used to convert an SR file into an MD.ai Annotation note or JSON format. It is meant to convert the entirety of the SR's content sequence into text while keeping note of the referenced studies.
The JSON format will have two fields: "Referenced DICOM" and "SR Content". Referenced DICOM is a list of dictionaries containing a "Study UID" field which points to the referenced studies (indicated by the content sequence's ReferencedSOPSequence tag). SR Content will be a list of lines of text, each containing a field from the SR document.
The annotation note format will import an annotation into the given project and dataset based on the studies that the SR references. The annotation will have a note containing the SR's content. You must initiate an mdai_client for this to work. Additionally, you must go into the UI and create a label, then input the label_id into the function.
Example:
import mdai
# Get variables from project info tab and user settings
DOMAIN = 'public.md.ai'
YOUR_PERSONAL_TOKEN = '8b80c4ca0f43421'
mdai_client = mdai.Client(domain=DOMAIN, access_token=YOUR_PERSONAL_TOKEN)
dataset_id = 'D_0Z4qeN'
project_id = 'L1NpnQBv'
label_id = 'L_QnlPAg'
from glob import glob
sr_files = glob("/path/to/dicom-sr/files/*.dcm")
mdai_client.import_sr(sr_files,
dataset_id=dataset_id,
project_id=project_id,
label_id=label_id)
Importing 15 annotations into project L1NpnQBv, dataset D_0Z4qeN...
Successfully imported 15 / 15 annotations into project L1NpnQBv, dataset D_0Z4qeN.
Export/Import notebook
This linked notebook gives examples for using the mdai library in order to:
- download annotations from your project
- turn the exported JSON file into a spreadsheet-like dataframe
- import/upload data or outside model predictions back into your project
You'll need to supply the specifics for your project, but the template in the notebook will get you started