MD.ai Interface Code
Consider the situation where you have trained your own model or have an existing model/implementation that you want to test on a dataset and visualize the predictions in a simple and user-friendly manner. MD.ai provides an efficient way to achieve this by simply uploading a zip file consisting of the model code arranged in a format that is compatible with our interface. This document summarizes the main files that are necessary for glueing your model with the MD.ai interface so that you can successfully run inference tasks without any hassle.
Inside the folder that contains your model weights file, you first need to create a folder named .mdai/. This folder will contain all the necessary files required for interacting with the interface. There are three main files that are needed in order to achieve this goal:
- config.yaml
- mdai_deploy.py
- requirements.txt
In addition to these files, you can also add helper python files that store methods for aiding the inference task, for example files that store preprocessing/postprocessing functions.
Explicit additions of folders to sys.path
We explicitly add all the folders within the zip file (that you upload to the md.ai interface) to sys.path. So you can add additional helper code files to any folder and this will allow you to import modules freely (even from within .mdai/).
To make things easier, you can download the zip file containing skeleton code for the MD.ai interface here and fill in the blanks as you read through the documentation here. Note that the folder .mdai inside the folder modelis a hidden folder and so you might not see it when you extract the zip file unless your system has the Show hidden files option on.
Let's go into the details of what goes inside the necessary files now.
config.yaml
The config.yaml file is the main configuration file that defines the runtime requirements for your model. As of now, it should contain the following tags:
- base_image: This tells Docker what to use as the base python environment image. Currently, we support
py37,py38,py39andpy310for setting up a python 3.7, 3.8, 3.9 or 3.10 conda environment respectively. (required) - device_type: This tells whether to run the model on a
cpuorgpu(default: cpu) - cuda_version: This tells which CUDA version to use in the python environment. This is required only if you have specified
gpuas the device type. Currently supported versions of CUDA include 10.0, 10.1, 11.0 and 11.3. (default: 11.0)
For example, a basic config file looks like the following -
We use Google cloud platform's cpu/gpu docker images (as parent images) to create derivative containers depending on what you select as the device_type. These container images provide us with a Python 3 environment and include Conda along with the NVIDIA stack for GPU images (eg. CUDA 11.0, cuDNN 7.x, NCCL 2.x). We build a conda environment on top of this and install all the necessary frameworks used by your model, reading from the requirements.txt which is explained later on this page.
mdai_deploy.py
The mdai_deploy.py contains the main python code that will be called for running the model on the given dataset and generating predictions. The main requirement for this file is to create a class named MDAIModel that will have two methods, namely:
- __init__: Defines the necessary values that will successfully initialize the model, for example path to the model checkpoint file and the model definition itself.
- predict: Contains the code for reading the input files, preprocessing them and passing them through the model to generate predictions. This method should return a list of dictionaries called
outputsthat will be read by the MD.ai interface.
Let's dig deeper into how to transform your code so that it fits the description of the MDAIModel class. But first, we need to understand the schema of the input data that is read from the interface and the schema of the outputs dict that will be returned.
Once you create a project and upload data on our interface, the data gets stored as msgpack-serialized binary data and has the following schema:
{
# image file
"files": [
{
"content": "bytes",
"content_type": "str", # MIME type, e.g. 'application/dicom'
"dicom_tags": "dict", # only available if input type is Thumbnail or when working with cloud storage, the images are JPEG/PNG
},
...
],
# annotations on md.ai for the image, if any
"annotations": [
{
"id": "str",
"label_id": "str",
"study_uid": "str",
"series_uid": "str",
"instance_uid": "str",
"frame_number": "int",
"data": "any",
"parent_id": "str"
},
...
],
# model label classes details
"label_classes": [
{
"class_index": "int",
"label": {
"id": "str",
"name": "str",
"type": "str", # 'GLOBAL', 'LOCAL'
"scope": "str", # 'INSTANCE', 'SERIES', 'STUDY'
"annotation_mode": "str", # For local annotation types
"short_name": "str",
"description": "str",
"parent_id": "str"
}
}
...
],
"args": {
"arg1": "str",
"arg2": "str",
...
}
}
The data["files"] will contain all the images within the specific batch in a bytes format. You can loop through this to access each file individually.
If during the model config setup on the UI (as will be explained in later sections), you select the Input file type to be DICOM, then the content type for each file in data["files"] will be file["content_type"] = 'application/dicom' and file["content"] is the raw binary data representing the DICOM file, which can be loaded using: ds = pydicom.dcmread(BytesIO(file["content"])) as we'll show in an example later.
If instead you select the Input file type to be Thumbnail or you are working with JPEG/PNG images that are stored in cloud storage datasets, then the content type will be file["content_type"] = 'image/jpeg' or 'image/png' as instead of raw DICOMs we will now be sending raw jpeg/png images to the model as input. For such cases we will additionally send the DICOM tags separately to the model which you can find within file["dicom_tags"].
Using uploaded JPEG/PNG images on MD.ai for models
If you upload JPEG/PNG images to a dataset on MD.ai and want to run models on them, kindly check out the Working with DICOM converted from JPEG/PNG section for additional preprocessing considerations.
If you have annotated the image on MD.ai, then the annotations for this specific input image can also be accessed under file["annotations"]. Note that annotations for only those labels that have been added as label classes to your model card on the UI, will be passed as inputs. You can also get the model label classes details for the particular model by accessing file["label_classes"].
Another thing to note is that once you create a new model version on your project (as explained in the Deploying models page) you have the option to specify the model scope for your model i.e does the model runs and produces outputs for each instance individually, or for a series or a study/exam as a whole. Model scope thus specifies whether an entire study, series, or instance is returned to the model class from the storage bucket.
- If the model scope is
INSTANCE, thenfileswill be a single instance (list length of 1). - If the model scope is
SERIES, thenfileswill be a list of all instances in a series. - If the model scope is
STUDY, thenfileswill be a list of all instances in a study.
If multi-frame instances are supported, the model scope must be SERIES or STUDY, because internally we treat these as DICOM series.
The additional args dictionary supplies values that may be used in a given run.
Now once the images are loaded this way, the next step is to add the necessary code for running this input through the model. It can simply be achieved by passing this input through already existing functions in helper files or you can explicitly add code inside the predict method to generate predictions.
Once the model returns an output, the predict method needs to return results in a particular schema (required for our interface to read and display outputs correctly) as shown below:
[
{
"type": "str", # choose from {'NONE', 'ANNOTATION', 'DICOM', 'IMAGE'}
"study_uid": "str",
"series_uid": "str",
"instance_uid": "str",
"frame_number": "int", # indexed from 0, only required when dealing with multiframe instances
"class_index": "int",
"data": {},
"probability": "float" or "list[dict[str, float]]",
"note": "str",
"explanations": [
{
"name": "str",
"description": "str",
"content": "bytes",
"content_type": "str",
},
...
],
},
...
]
type defines the type of output that is produced, whether it is an annotation, or DICOM image, png/jpeg image or NONE type (needs to be entered explicitly).
study_uid defines the unique ID of the study to which the particular instance belongs (present in the instance DICOM tags).
series_uid defines the unique ID of the series to which the particular instance belongs (present in the instance DICOM tags).
instance_uid defines the unique ID of the particular instance (present in the instance DICOM tags).
frame_number defines the frame number for multiframe instances. When dealing with multiframe, MD.ai treats each frame as a single image and so the frame number helps map outputs to the corresponding frame. Frame numbers are indexed from 0.
class_index defines the output class according to your model definition for the particular instance and should map to the labels created on the MD.ai interface.
data defines a dictionary of resulting annotations such as bounding box coordinates. (optional)
note defines any textual description that you can assign to each model output. This note will be displayed in the separate notes panel as well. (optional)
probability defines the probability of the output belonging to the specified class_index if your model produces a probability value. For multiclass and multilabel outputs you should return a list of dictionaries that contains all class probabilites along with their respective class indices - [{"class_index": 0, "probability": 0.1}, ..., {"class_index": n, "probability": 0.9}]. (optional)
explanations define additional exploratory studies such as GradCAM, SmoothGrad analysis or any other instance related results that you want to display. Check out adding a heatmap (GradCAM) as an explanation here (optional)
Note
The DICOM UIDs must be supplied based on the scope of the label attached to class_index.
An example file might look like the following -
# Import statements for the python packages used
# Import methods from helper files if any
# Create an MDAIModel class with __init__ and predict methods
class MDAIModel:
def __init__(self):
modelpath = # Path to your model checkpoint file
# examples
# modelpath = os.path.join(os.path.dirname(os.path.dirname(__file__)), "model_file.pth")
# modelpath = os.path.join(os.path.dirname(os.path.dirname(__file__)), "model_file.h5")
self.model = # Load the model file
def predict(self, data):
# Load the input files
input_files = data["files"]
# Load the input data arguments (if any)
input_args = data["args"]
# Load human annotations as input (if any)
input_annotations = data["annotations"]
# List for storing results for each instance
outputs = []
# Loop through the data points
for file in input_files:
# Check if the file type is dicom or any other format.
if file['content_type'] != 'application/dicom':
continue
# Read the dicom file (if using pydicom)
ds = pydicom.dcmread(BytesIO(file["content"]))
# Convert dicom to a numpy array of pixels
image = ds.pixel_array
# Code for preprocessing the image
# Code for passing the image through the model
# and generating predictions
# Store results in a dict following the schema mentioned, for example -
result = {
"type": "ANNOTATION",
"study_uid": str(ds.StudyInstanceUID),
"series_uid": str(ds.SeriesInstanceUID),
"instance_uid": str(ds.SOPInstanceUID),
"class_index": int(class_index),
"data": {},
"probability": float(probability) or [{"class_index": int(class_index), "probability": float(probability)}, ...],
"explanations": [
{
# Add explanations if any
},
],
}
# Add results to the list
outputs.append(result)
# Return list to be read by the MD.ai interface
return outputs
Make sure to type cast the returned values as shown in the example above. This example file can also be used as skeleton code which you can edit according to your needs and requirements. For more examples, check out the code for our X-ray classification model and Lung segmentation model which we have already deployed on our platform for reference.
requirements.txt
In order for your model to run successfully on our interface, it is important that the correct versions of the python packages used for running your model be installed inside our environment. These packages are mostly those you import in the mdai_deploy.py file along with those in any helper files that you use. These can be easily installed by simply creating a text file by the name requirements.txt that contains the package name along with the specific version that needs to be installed.
An example requirements.txt file looks like this:
or to add a git library from source like detectron2 use:
or to add libraries that use the --find-links or -f option with pip install, add the link to archives such as sdist (.tar.gz) or wheel (.whl) files in one line, followed by the package name in the next line. For example, pre-built detectron2 that uses torch 1.6 and CUDA 10.1 can be installed by adding the following two lines to the requirements.txt file:
or to add custom .whl files for installation, add all .whl files alongside the requirements.txt file within the .mdai folder of your zip file. Then, reference these .whl files for installation in your requirements.txt file as follows -
Note
In order to find the correct versions of the required packages, run PACKAGE.__version__ to check what version you are using locally.
We provide the following packages, with their corresponding versions, pre-installed. You can save build time by not mentioning these in your requirements.txt file if the versions match your dependency requirements:
fastapi==0.82.0
msgpack==1.0.4
uvicorn==0.18.3
numpy==1.23.3
pydicom==2.3.0
pylibjpeg==1.4.0
pylibjpeg-libjpeg==1.3.1
pylibjpeg-openjpeg==1.2.1
pylibjpeg-rle==1.3.0
protobuf==3.20.1
Once these files are ready, store them in a folder named .mdai and follow the next steps as mentioned on the page Deploying models.
Supported model output label types
Our annotator supports creation of both global and local level labels. Global labels include Exam, Series and Instance scoped labels. Local labels are instance scoped by default and include Mask, Freeform, Bounding Box, Polygon, Line and Location types. You can read more details about label creation here.
All of these annotation types are thus supported for deploying models and you can easily deploy your choice of classification, segmentation, object detection model using this to name a few.
Based on the kind of model and the outputs associated, we require the returned list of dictionaries (outputs in the example above) in the mdai_deploy.py file to include all outputs in a format that is compatible with our interface.
Global Labels
For global classification labels, based on the scope chosen for the output label on our interface interface you need to return the following -
result = {
"type": "ANNOTATION",
"study_uid": str(ds.StudyInstanceUID), # required for all EXAM, SERIES and INSTANCE scoped labels
"series_uid": str(ds.SeriesInstanceUID), # required for SERIES and INSTANCE scoped labels
"instance_uid": str(ds.SOPInstanceUID), # required for INSTANCE scoped labels
"class_index": int(class_index), # required for all labels
"probability": float(probability) or [{"class_index": int(class_index), "probability": float(probability)}, ...], # optional
"explanations": [
{
# optional, add explanations if any such as gradcam images
},
],
}
Local Labels
For local labels, all the UIDs are required to be returned along with the the data key that tells what type of output it is among mask, bounding box, freeform, polygon, line and location. The following needs to be returned -
result = {
"type": "ANNOTATION",
"study_uid": str(ds.StudyInstanceUID), # required for all labels
"series_uid": str(ds.SeriesInstanceUID), # required for al labels
"instance_uid": str(ds.SOPInstanceUID), # required for all labels
"class_index": int(class_index), # required for all labels
"data": {}, # required based on the label type
"probability": float(probability) or or [{"class_index": int(class_index), "probability": float(probability)}, ...], # optional
"explanations": [
{
# optional, add explanations if any such as gradcam images
},
],
}
Warning
Make sure you return the right format for data depending on the type of label you create on the interface. For example, creating a freeform label on our interface and returning the data in the mask format will not work.
For the data key -
-
Mask: If the output is a segmentation mask, you need to create a
Masklabel on the interface and return the data as -"data": {"mask": []}Note that the mask needs to be returned as a list. If your mask is produced as a numpy array, you need to convert it to a list using the
.tolist()method, eg."data": {"mask": your_mask.tolist()} -
Bounding box: If the output is a bounding box, you need to create a
Bounding Boxlabel on the interface and return the data as -"data": {"x": int, "y": int, "width": int, "height": int}where
xandyare the coordinates of the top left corner of your bounding box. -
Freeform, Polygon or Line: If the output is a freeform shape, polygon or line, you need to create the appropriate label on the interface and return the data as -
"data": {"vertices": [[[0,0],[0,0]], [[1,1], [1,1]]}Note that
verticesneeds to be a list of lists where each sublist contains the (x,y) coordinate of a point. You can use the scikit-image library to convert your outputs into contours for this. -
Location: If the output is a point, you need to create a
Locationlabel on the interface and return the data as -"data": {"x": int, "y": int}where
xandyare the coordinates for the point.
DICOM/IMAGE type outputs
It is also possible to return DICOM or JPEG/PNG images as model outputs (for example images generated using generative AI models like stable diffusion) and view them in the MD.ai viewer. There is also support to export these generated images to a new dataset in the same project.
If your model generates JPEG/PNG images or images in the form of numpy arrays -
You need to set type: "IMAGE" in your output dictionary and save the generated image as a BytesIO() object before returning it. The generated image goes into the key named "images".
To return a single generated image, follow this code snippet for the required lines to add:
# Image generated by the model (numpy array or existing png image)
generated_image = PIL.Image.fromarray(image_array) or PIL.Image.open("test.png")
# save image as a buffer
image_buffer = BytesIO()
generated_image.save(image_buffer, format="PNG") # or jpg
outputs.append(
{
"type": "IMAGE",
"study_uid": str(ds.StudyInstanceUID),
"series_uid": str(ds.SeriesInstanceUID),
"instance_uid": str(ds.SOPInstanceUID),
"class_index": 0,
"note": "This is a generated image",
"images": [image_buffer.get_value()], # return image encapsulated in a list
}
)
Once the model runs successfully and returns the generated image, you can view the image by pressing the Open Image in Viewer button on the right of the model output.
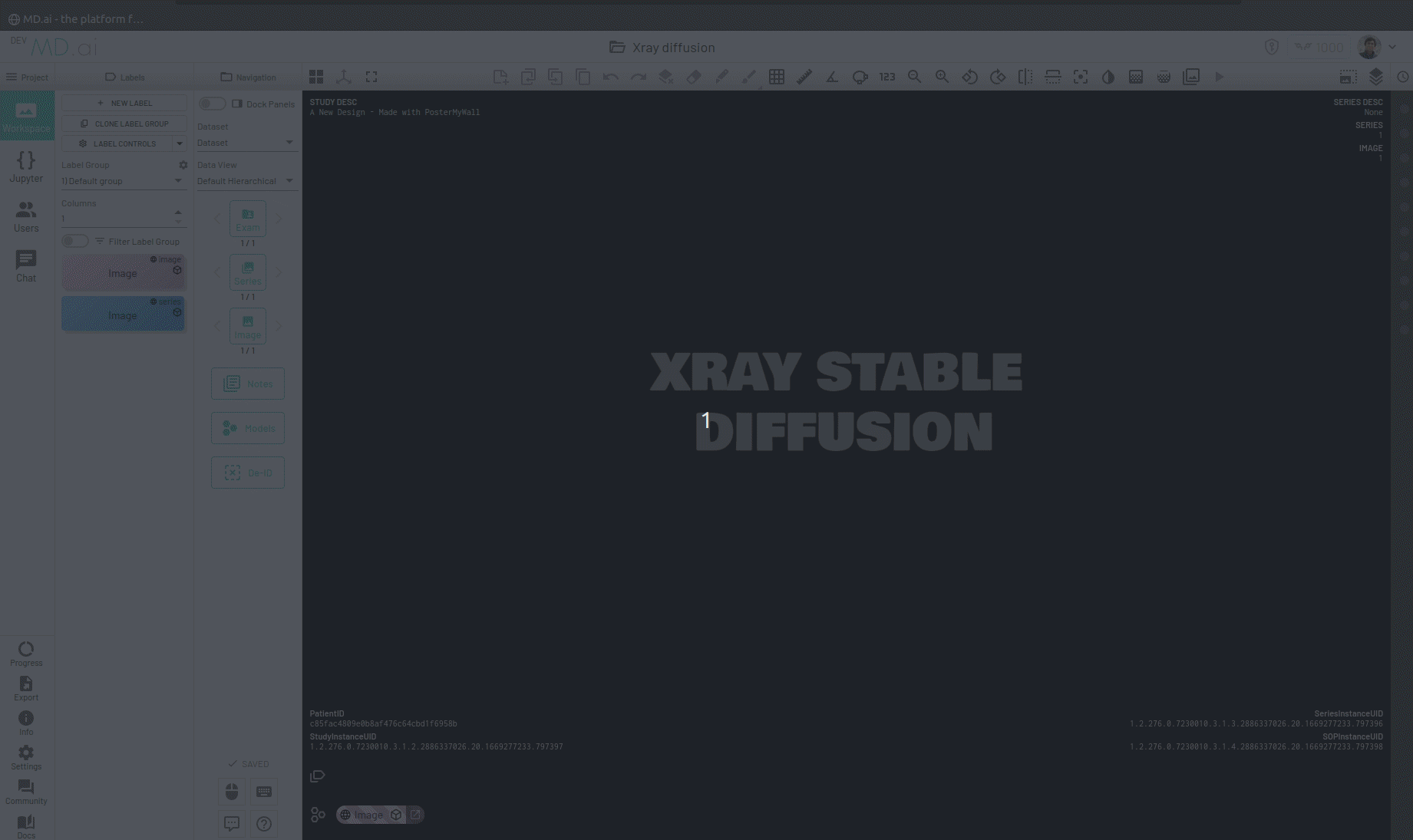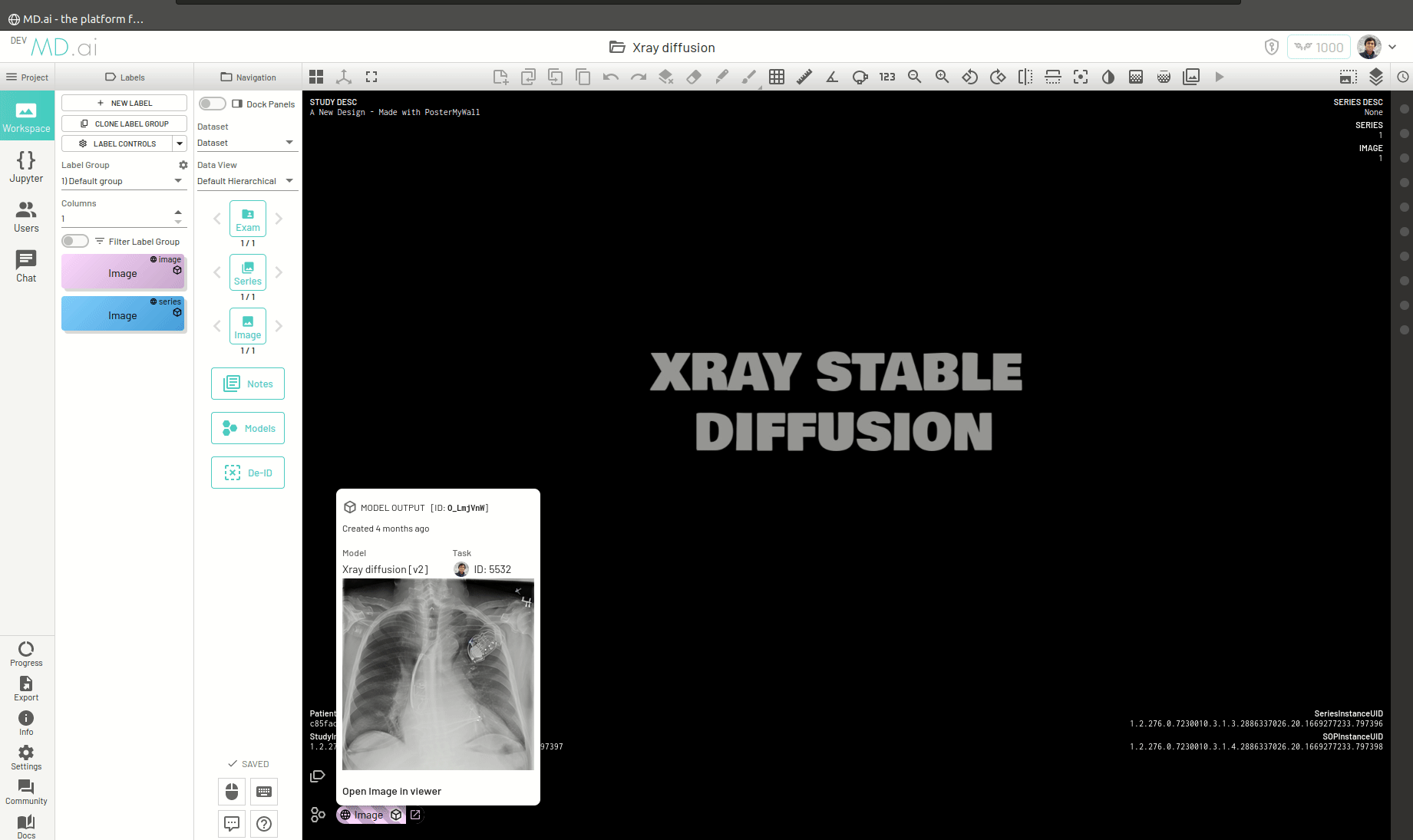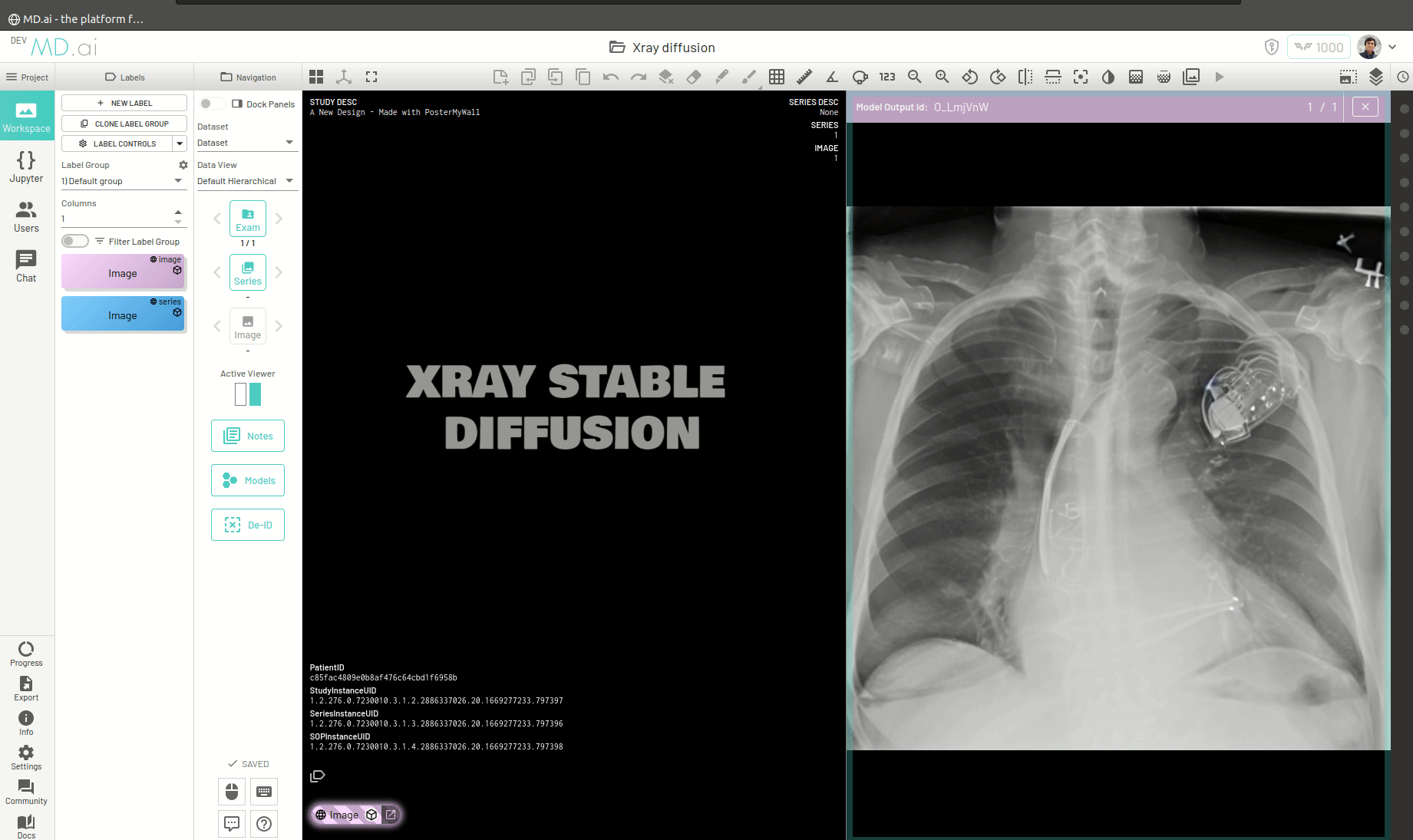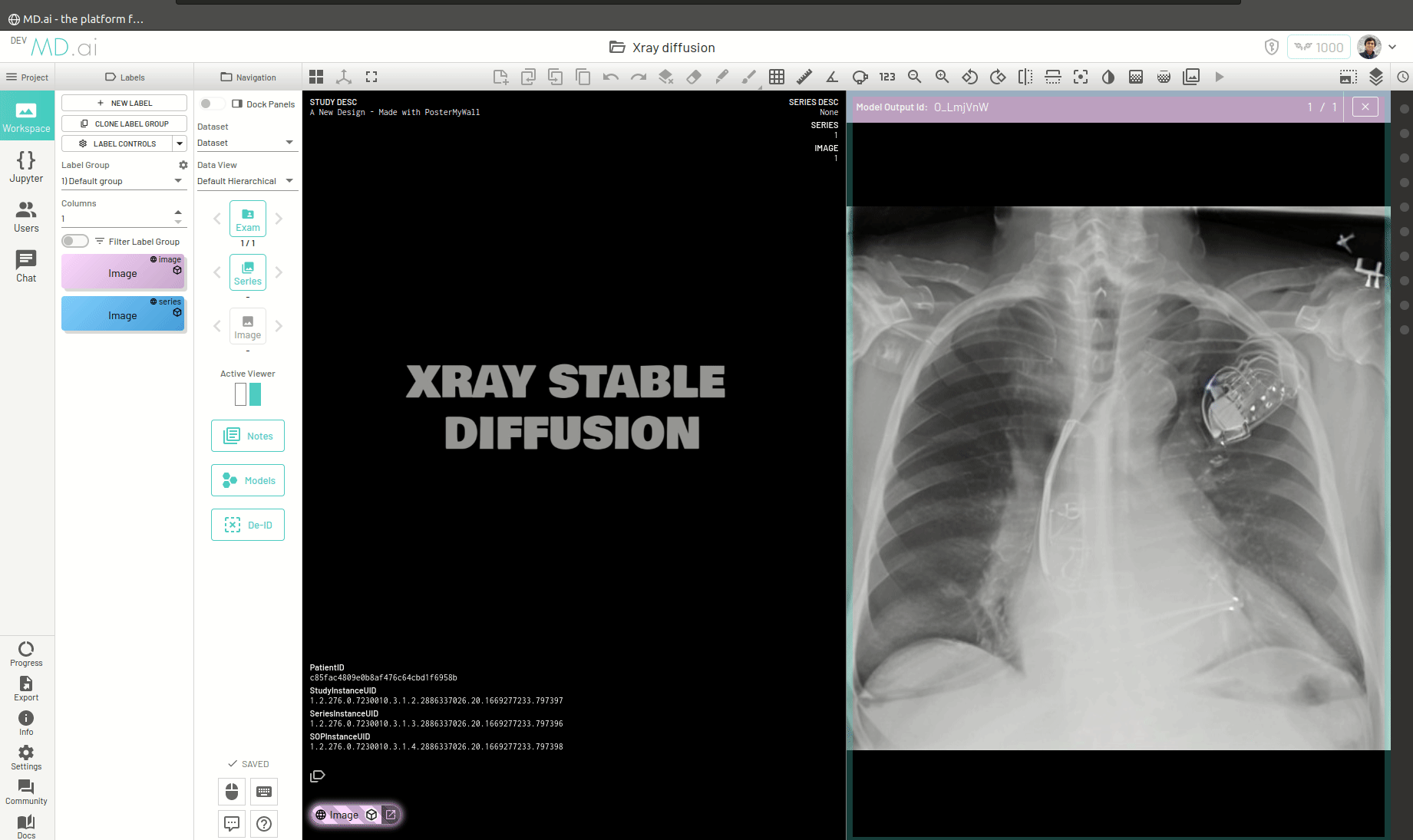
You can also return a bunch of generated images as a series (for example when generating volumetric 3D images like CTs). For this you need to add all the generated images in a list and return that list as the value for the images key in the output dictionary. For example -
# list of images generated by the model
generated_images = [x, y, z]
# loop through each generated image and save it as a BytesIO() object
ims = []
for j, i in enumerate(generated_images):
image_buffer = BytesIO()
i.save(image_buffer, format="PNG")
ims.append(image_buffer.getvalue())
outputs.append(
{
"type": "IMAGE",
"study_uid": str(ds.StudyInstanceUID),
"series_uid": str(ds.SeriesInstanceUID),
"instance_uid": str(ds.SOPInstanceUID)
"class_index": 0,
"note": "This is a generated image series",
"images": ims,
}
)
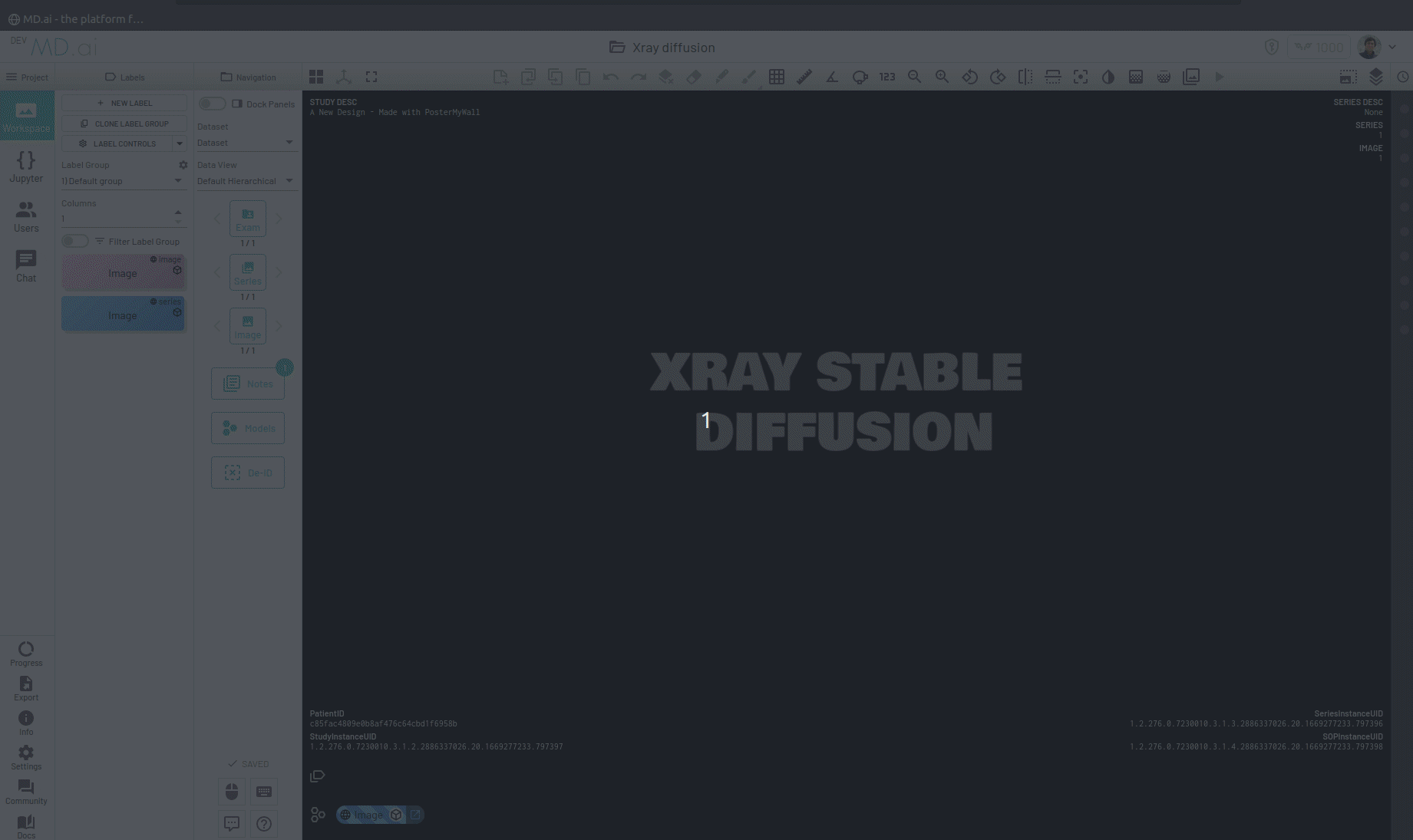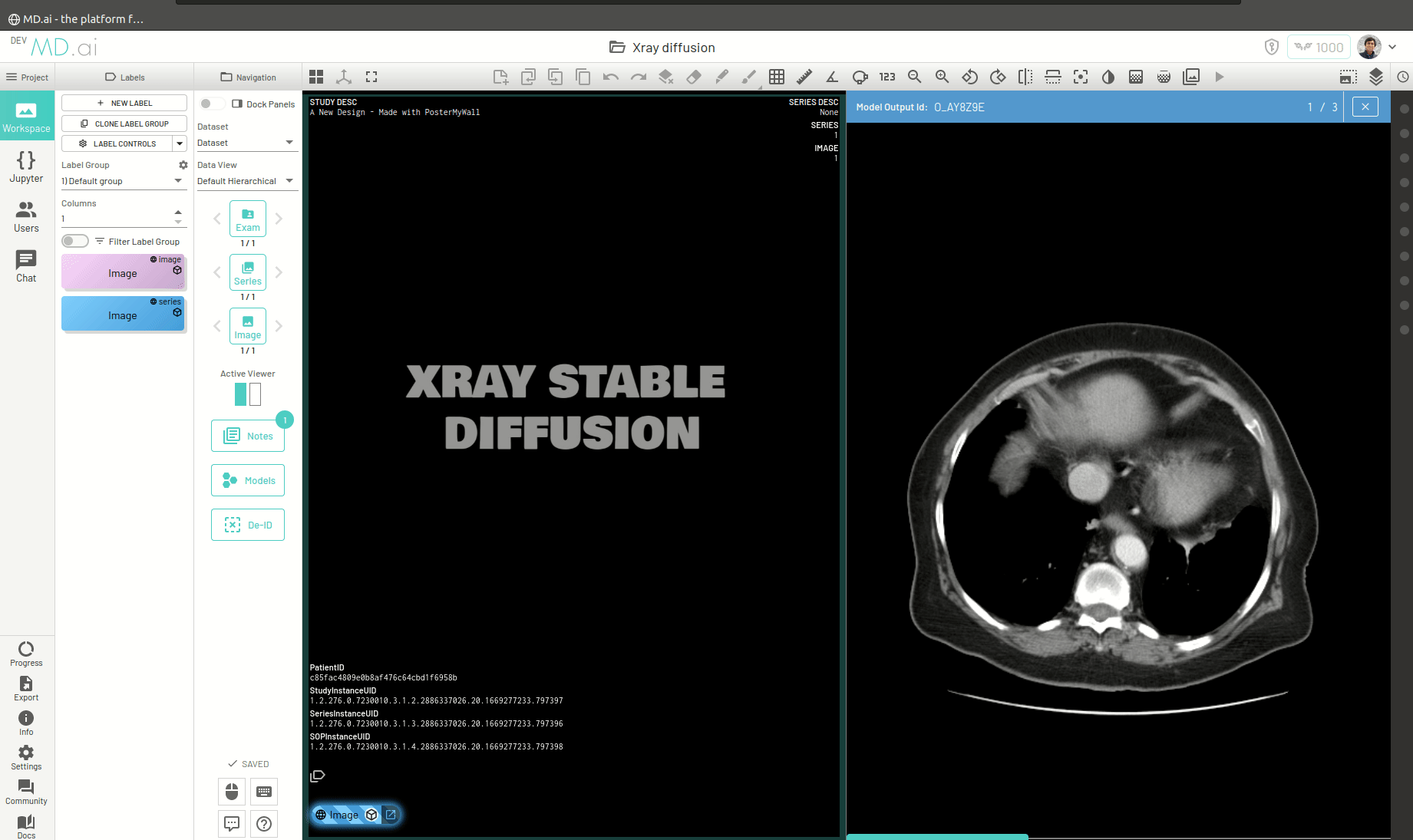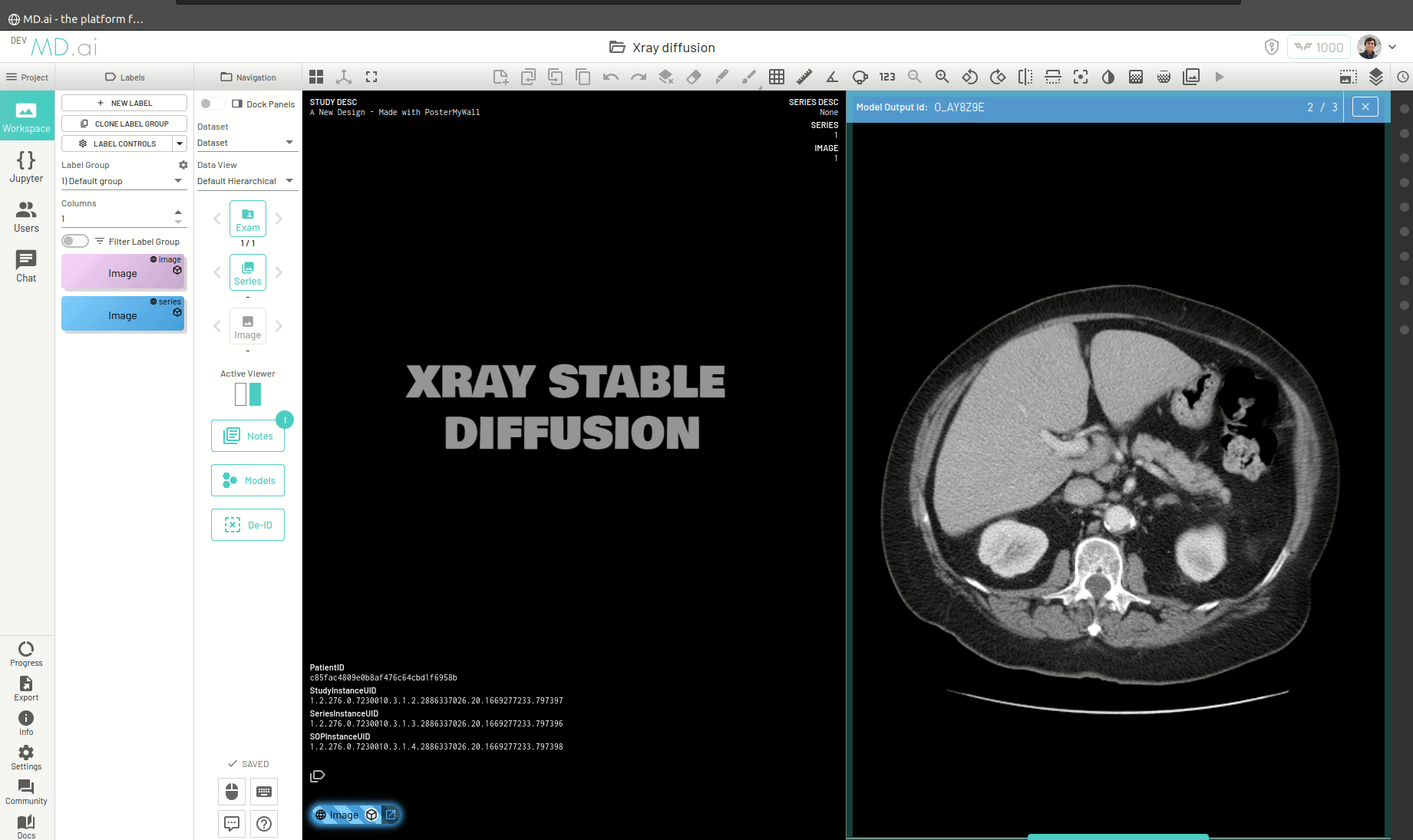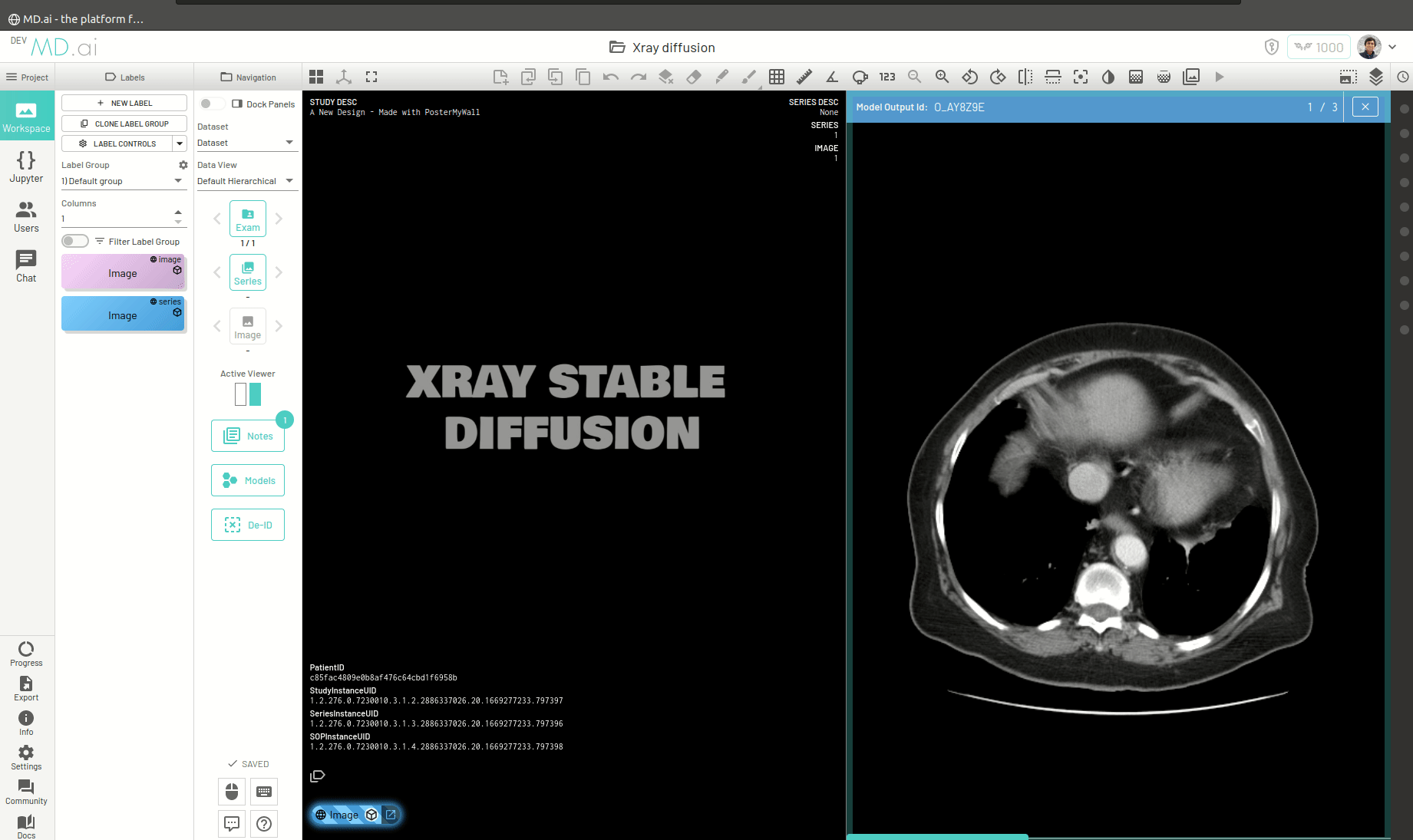
When converting such IMAGE type outputs to a new dataset, you can also choose to pass certain DICOM tags that will be preserved or fixed during the conversion. Currently we support passing the PatientID, StudyInstanceUID and SeriesInstanceUID tags. For example for the above example series, you can fix these tags by adding a new key called image_output_tags in the returned output dictionary:
outputs.append(
{
"type": "IMAGE",
"study_uid": str(ds.StudyInstanceUID),
"series_uid": str(ds.SeriesInstanceUID),
"instance_uid": str(ds.SOPInstanceUID)
"class_index": 0,
"note": "This is a generated image series",
"images": ims,
"image_output_tags": {"PatientID": "test", "StudyInstanceUID": "1.2.826.0.1.3680043.8.498.10103161217531758106941925783974775366", "SeriesInstanceUID": "1.2.826.0.1.3680043.8.498.43206709821313539967892922635326990285"} # The new converted DICOM will preserve these tags
}
)
If your model generates DICOM images -
The steps required are the same as before for the Image type model, but you just need to use type: "DICOM" while returning dicoms.
# Image generated by the model (numpy array or existing png image)
generated_dicom = pydicom.dcmread(image_array)
image_buffer = BytesIO()
generated_dicom.save_as(image_buffer)
# save image as a buffer
image_buffer = BytesIO()
generated_image.save(image_buffer, format="PNG") # or jpg
outputs.append(
{
"type": "IMAGE",
"study_uid": str(ds.StudyInstanceUID),
"series_uid": str(ds.SeriesInstanceUID),
"instance_uid": str(ds.SOPInstanceUID),
"class_index": 0,
"note": "This is a generated image",
"images": [image_buffer.get_value()], # return image encapsulated in a list
}
)
For returning a bunch of DICOMs as a series, add them to a list and return them. The order of the files in the list is again important to determine how the images will be arranged in the series (instance number).
Note
Note that since on MD.ai the outputs are always associated with an input image, the study_uid, series_uid and instance_uid correspond to the INPUT dicom image/resource that you are running the model on. They should not be confused with the values for the generated DICOM files.